20 November 2025 – A new pharmacogenomics initiative in Iran provides population-specific data for 168 clinically relevant drugs. By characterising allele frequencies across 37 genes, it aims to reduce adverse drug reactions and optimise treatments in oncology, cardiology and psychiatry. The Tehran University-led project aligns with CPIC standards for clinical implementation.
Benefit for Personalised Medicine
Healthcare is increasingly shifting from a one-size-fits-all approach to a more personalised focus on individual patients. Personalised medicine is an emerging approach that focuses on identifying specific targets for treatment and related diagnostic tools, to improve the efficacy and safety of medical interventions. Pharmacogenomics (PGx) studies the inter-individual variations of different genes to variable drug response with the ultimate goal of improving therapeutic approaches. In addition, the incorporation of pharmacogenomics into clinical practice has gained broader acceptance, with its increasing application in the identification of novel drug targets and the development of drugs tailored to specific patient groups within the framework of precision medicine. Key benefits of pharmacogenomics for personalised medicine are optimised drug selection, reduced adverse drug reactions, improved drug dosing, increased treatment efficacy, cost-effectiveness, empowered clinical decision-making, facilitation of preventive care. While pharmacogenomic screening has been established in numerous medical facilities in the United States and Europe, its adoption in other regions of the world, such as the Middle East, is lagging primarily due to insufficient data availability.
The pharmacogenomics initiative in Iran
The study is designed in three major axes genotyping approach, gene Selection based on CPIC guidelines, and data analysis. The main objective is to characterise allele frequencies of 1040 variants across 37 key pharmacogenes, guided by the Clinical Pharmacogenetics Implementation Consortium (CPIC).
29 Drug Categories = 168 Drugs
- CFTR and ivacaftor
- CYP2B6 and efavirenz
- CYP2B6 and Methadone
- CYP2C19 and Clopidogrel (Plavix®)
- CYP2C19 and Proton Pump Inhibitors
Dexlansoprazole, Esomeprazole, Lansoprazole, Omeprazole, Pantoprazole and Rabeprazole - CYP2C19 and Voriconazole
- CYP2C9 and Non-steroidal anti-inflammatory drugs (NSAIDs)
Aceclofenac, Celecoxib, Diclofenac, Flurbiprofen, Ibuprofen, Indomethacin, Lornoxicam, Lumiracoxib, Meloxicam, Nabumetone, Naproxen, Piroxicam and Tenoxicam - CYP2C9, HLA-B and Phenytoin
Fosphenytoin and Phenytoin - CYP2C9, VKORC1, CYP4F2 and Warfarin
- CYP2D6 and Ondansetron and Tropisetron
Ondansetron and Tropisetron - CYP2D6 and Atomoxetine
- CYP2D6, ADRB1, ADRB2, ADRA2C, GRK4, GRK5 and Beta-Blockers
Acebutolol, Atenolol, Betaxolol, Bisoprolol, Carvedilol, Esmolol, Labetalol, Metoprolol, Nadolol, Nebivolol, Pindolol, Propranolol and Sotalol - CYP3A5 and Tacrolimus
- CYP2D6, CYP2C19, CYP2B6, SLC6A4, HTR2A and Selective Serotonin Reuptake Inhibitors (SSRIs)
Citalopram, Duloxetine, Escitalopram, Fluoxetine, Fluvoxamine, Paroxetine, Sertraline, Venlafaxine and Vortioxetine - CYP2D6 and Tamoxifen
- CYP2D6, OPRM1, COMT and Opioids
Codeine, Hydrocodone, Oxycodone, Tramadol and Acetaminophen/caffeine/dihydrocodone
- G6PD
Aminosalicylic acid, Aspirin, Chloramphenicol, Chloroquine, Chlorpropamide, Ciprofloxacin, Dabrafenib, Dapsone, Dimercaprol, Doxorubicin, Furazolidone, Gliclazide, Glimepiride, Glipizide, Glyburide, Hydroxychloroquine, Mafenide, Mepacrine, Mesalazine, Methylene blue - TPMT, NUDT15 and Thiopurines
Azathioprine, Mercaptopurine and Thioguanine - DPYD and Fluoropyrimidines
Cepecitabine, Fluorouracil and Tegafur - SLCO1B1, ABCG2, CYP2C9 and Statins
Atorvastatin, Fluvastatin, Lovastatin, Pitavastatin, Pravastatin, Rosuvastatin and Simvastatin - UGT1A1 and Irinotecan
- CYP2D6, CYP2C19 and Tricyclic Antidepressants (TCAs)
Amitriptyline, Clomipramine, Desipramine, Doxepin, Imipramine, Nortriptyline and Trimipramine - HLA-A, HLA-B and Carbamazepine and Oxcarbazepine
Carbamazepine and Oxcarbazepine - HLA-B and Abacavir
- HLA-B and Allopurinol
- IFNL3 and Peginterferon-alpha-based Regimens
Pegintereron alfa-2, Peginterferon alfa-2b. Ribavirin - MT-RNR1 and AminoglycosidesAmikacin, Dibekacin, Gentamicin, Kanamycin, Neomycin, Netilmicin, Paromomycin, Plazomicin, Ribostamycin, Streptomycin, Tobramycin
- RYR1, CACNA1S and Volatile anesthetic agents and Succinylcholine
Desflurane, Enflurane, Halothane, Isoflurane, Methoxyflurane, Sevoflurane, Succinylcholine - UGT1A1 and Atazanavir
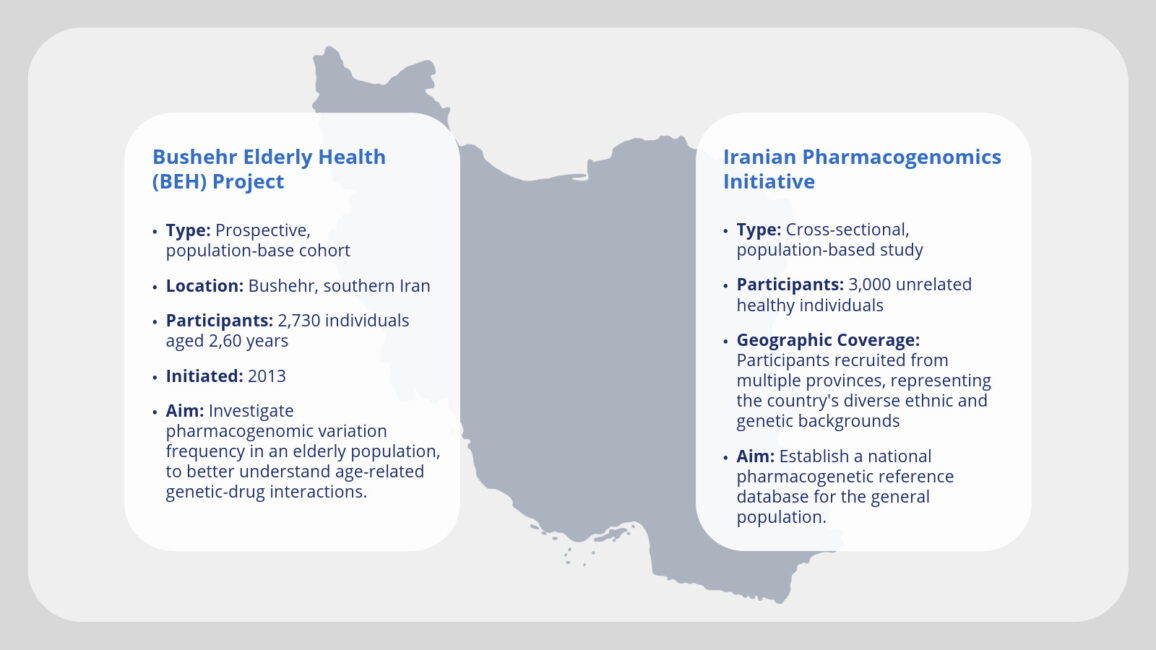
This initiative aligns with reporting guidelines for precision medicine research of clinical relevance: the BePRECISE checklist. The BePRECISE guidelines, developed by a global consortium of experts, provide a framework for reporting precision medicine research to improve the quality, comparability, and reproducibility of studies. These guidelines address the need for standardised reporting in precision medicine, which has been hampered by the lack of such standards. The BePRECISE Checklist, comprising 23 items organised into five sections, aims to enhance the accuracy, safety, and health equity in precision medicine research. Two independent study cohorts are considered:
Strategic Integration
Based on the current progress of the initiative, we are entering the early stages of clinical implementation, particularly in therapeutic areas where pharmacogenomic evidence is strongest such as oncology, cardiology, and psychiatry. This transition from research to clinical utility is now supported by national policy and governmental endorsement. Most notably, on April 14, 2025, the Iranian Ministry of Health and Education officially launched the “Personalised Medicine Ecosystem” and unveiled the Pharmacogenomics initiative results in a high-level event hosted at Tehran University of Medical Sciences. The event was attended by top medical officials and academic leaders. This policy endorsement aligns with the ongoing efforts of the Iranian Pharmacogenomics Initiative, facilitating the translation of genomic research into clinical practice. The government’s support underscores the importance of developing infrastructure, regulatory frameworks, and educational programs to ensure the effective implementation of pharmacogenomic testing and personalised treatment strategies.
Future perspective
The initiative is poised to move into several key next-phase priorities over the upcoming months and years. These steps go beyond infrastructure and guideline alignment and focus on education, clinical translation, national scaling, and regional leadership.
Future steps are:
- Development of National Clinical Guidelines
- Building on the CPIC framework: Collaboration with medical associations to mandate or recommend PGx testing for specific high-risk medications.
- Training and Certification Programmes
- To ensure successful clinical rollout: Create certification courses for healthcare providers (clinicians, pharmacists, genetic counsellors) on how to interpret and apply pharmacogenomic data. Also it needs to integrate PGx education into medical and pharmacy school curricula.
Benefits of the pharmacogenomics initiative for patients
1. Foundation for Clinical Implementation
By identifying the allele frequencies of 1040 variants across 37 clinically actionable pharmacogenes (as defined by CPIC), this initiative is building the genetic reference database necessary for safe and effective drug prescribing in the Iranian population. This national reference database (http://irpkb.ir/) addresses a critical gap: most current clinical pharmacogenomics guidelines (e.g., CPIC) are based on allele frequency data derived from non-Iranian populations. Applying those directly to Iranian patients without our population validation could result in suboptimal or even unsafe pharmacogenetics recommendations. Our initiative ensures that genotype-guided prescribing in Iran will be based on population-relevant data, which improves both efficacy and safety. This directly enables future clinical decisions to be made with Iranian genomic data, which is more accurate than relying on international averages.
2. Identification of PGx Phenotypes
Preliminary analyses can already identify populations at risk of adverse drug reactions or treatment failure, such as poor metabolisers of CYP2D6 (antidepressants, opioids, etc.) or carriers of DPYD variants (risk for toxicity with fluoropyrimidines). A poor metaboliser is someone whose body can’t properly break down certain medications due to genetic differences. For example, CYP2D6 is a gene involved in processing many drugs like antidepressants, opioids, and tamoxifen. If someone has a non-working version of this gene, the medication might not work or could cause side effects. DPYD is a gene needed to safely break down chemotherapy drugs like 5-fluorouracil (5-FU). People with certain DPYD variants can experience severe, even life-threatening toxicity if treated without dose adjustment. The importance of DPYD testing is now officially recognised by major clinical guidelines. Notably, the 2025 update of the National Comprehensive Cancer Network (NCCN) guidelines for gastrointestinal cancers makes DPYD genotyping mandatory before initiating 5-FU-based chemotherapy. This represents a major step in integrating pharmacogenomics into routine oncology practice.
3. Population-Specific Drug Guidelines
Rather than relying solely on international databases (which often lack representation from Middle Eastern populations), we are now equipped to develop customised pharmacogenetic-based (PGx) prescribing guidelines tailored specifically to Iranian patients. These guidelines will adapt the clinical recommendations from international bodies like CPIC and DPWG, but incorporate Iranian allele frequencies to improve clinical validity and safety.
4. Education and Awareness
We are actively taking steps to raise awareness and build capacity among healthcare providers and researchers regarding the role of pharmacogenomics in personalised medicine. We have implemented educational workshops and CME programs, integration of pharmacogenomics into medical and pharmacy school curricula, establishment of personalised medicine network in the country by approval of Iranian ministry of health and education, and publications (“A comprehensive analysis across an ancestrally diverse Iranian population” and “Allele frequency of genetic variations related to the UGT1A1 gene”). Through these activities and the present study, healthcare providers and researchers are becoming more familiar with the role of pharmacogenomics in personalised medicine. This fosters a growing clinical and academic infrastructure that will support the integration of pharmacogenetic testing into routine care.
5. Policy and Infrastructure Development
Population-based pharmacogenomics dataset positions Iran to:
- Influence regional policy
- Attract international collaboration
- Establish infrastructure for pharmacogenomic clinical services, such as database of Iranian Pharmacogenomics Knowledge Base (IRPKB) (http://irpkb.ir/)
Mandana Hasanzad, PhD
Professor of Genetics
Personalized Medicine Research Center, Endocrinology and Metabolism Clinical Sciences Institute, Tehran University of Medical Sciences, Tehran, Iran.
Disclaimer: The views and opinions expressed are those of the author(s) only and do not necessarily reflect those of the European Partnership for Personalised Medicine (EP PerMed), national/regional funding organisations, the European Union or the European Commission. Neither of these parties can be held responsible for them.
